
PATIENCE, dung beetle!
About 35 years ago a ryegrass population that had been sprayed several times with Hoegrass (Diclofop) became resistant to that herbicide and cross resistant to Glean (chlorsulfuron) before Glean or any other ALS herbicide had ever been used in Australia.
P450 enzymes were suspected to be the cause of this cross resistance but it has taken until now to get the definitive evidence.
A very patient group of researchers led by Heping Han from the Australian Herbicide Resistance Initiative (AHRI), including researchers from Bayer and Zheijiang University in China have identified the P450 gene responsible for cross resistance to herbicides of at least five modes of action.
You’re going to love the name of the gene, it’s called: CYP81A10v7.
CYP is short for Cytochrome P450, a superfamily of enzymes. Plants contain hundreds of different P450 enzymes, so to find the specific one that causes this cross resistance is like finding a needle in a stack of needles! Hence the need for extreme patience, high level science, and an international team of molecular biologists.
In simple terms it involves turning up the dimmer switch to full noise on the production of the enzyme so the resistant plants have truckloads of the P450 enzyme that can break down the herbicide before they get to their site of action, or gene overexpression if you want to be technical.
The ryegrass population
This research was conducted on a famous ryegrass population called SLR31 that was sampled from Bordertown in South Australia in 1988, the same population that AHRI’s Roberto Busi recently used to ‘breed’ Sakura, Arcade and Avadex resistant ryegrass.
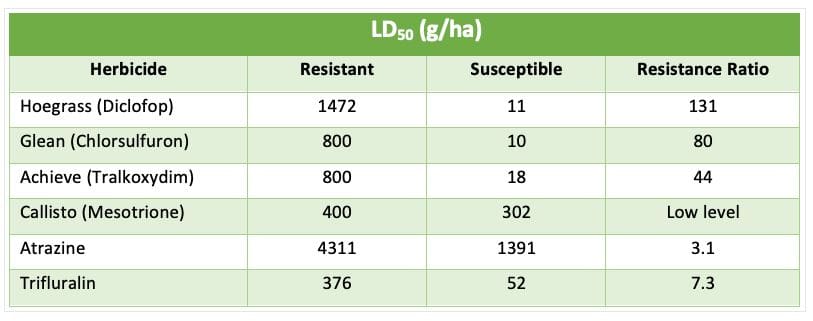
Table 1 below demonstrates what a cross resistant monster SLR31 is.
Table 1: LD50 (lethal dose to control 50% of the population) and resistance ratio for a range of herbicides for the control of Resistant (SLR31) and susceptible annual ryegrass.
Finding the gene
This is very high-level science that is difficult to understand, but here goes!
Step 1: Clone the ryegrass. This involves growing some ryegrass with many tillers then breaking the tillers off the plant to form a number of clones.
Step 2: Test the clones with herbicide treatment plus gene sequencing to identify the most resistant plants (ensuring that surviving clones do not have target-sits resistance). Now we have plants that are genetically similar but differing in herbicide resistance to take to step 3.
Step 3: Use RNA-Seq to identify a short list of genes that may be responsible for the resistance. RNA-Seq involves smashing the RNA into little pieces, reading the little pieces, then using a powerful computer to assemble the jigsaw puzzle and read the genome. From this they identified 8 candidates (called contigs) and three of them are P450 genes
At this stage we have found 8 needles in the needle stack, but which one is it?
Step 4: Use quantitative PCR (Q-PCR) to more accurately measure the genes contained on the strand of RNA. Q-PCR keeps the RNA strand whole and gives a more accurate measure of the level of gene expression.
Q-PCR found more than 500 times as much of the CYP81A10v7 gene expressed in the resistant plant as the susceptible. The other candidates had higher gene expression (2 to 16-fold), but nothing like the 500-fold expression of CYP81A10v7
Bingo! They found the needle in the stack of needles.
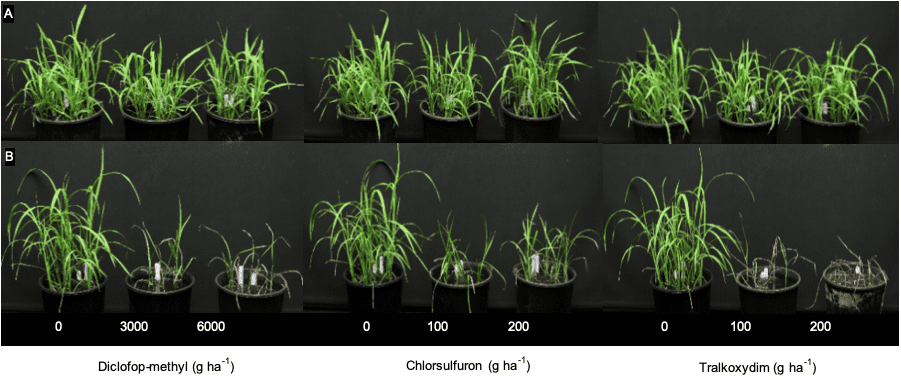

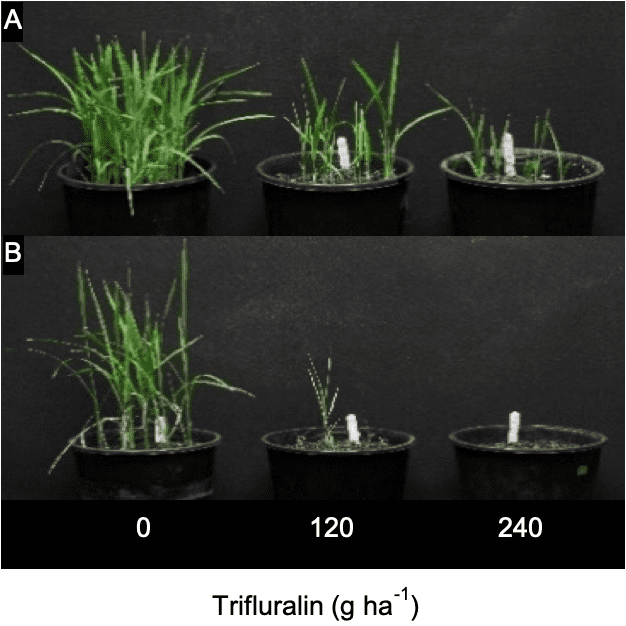
Rice calli and seedlings
To be extra sure, the researchers then created some rice calli (rice tissue grown on agar) and expressed the identified Lolium CYP81A10v7 gene in these calli. Rice seedlings generated from these transgenic calli were then tested with a range of herbicides. This confirmed that the over expression of this gene led to resistance of at least five herbicide modes of action.
What’s in a name?
CYP = Cytochrome P450
81A = the family (81) and subfamily (A). There are thousands of P450’s so they are broken up into families
10 = one of the children of that family
v7 = variant 7 of that child – ok, that is a bit weird but you get my drift
Summary
This is exciting research. There has been an international effort going for several years to truly understand non-target site metabolic resistance, and this research is a critical part of this understanding. This research is the culmination of 35 years of work and will be pivotal in understanding the cross-resistance patterns of our weeds and help us understand the risk of cross resistance to new herbicides.
Source: AHRI
Acknowledgements
This research is the fruit of great collaboration between AHRI and Bayer in Germany and financially supported by ARC and GRDC, with involvement of one of our AHRI Chinese scholars.
Authors
Heping Han, Qin Yu and Stephen B Powles, AHRI, University of Western Australia
Roland Beffa and Susana Gonzalez, Bayer AG, Weed Resistance Competence Center, Frankfurt, Germany
Frank Maiwald, Bayer AG, Monheim, Germany
Jian Wang, Zhejiang University, Hangzhou, China

HAVE YOUR SAY